A summary of my recent paper in Avian Research.
Traditionally, hybridization has been studied by comparing species pairs. But what happens when multiple species are interbreeding? This question led me to write a review on multispecies hybridization in birds, which was recently published in the journal Avian Research. In this blog post, I will walk you through the review and highlight some interesting results and discussion points. But I encourage you to read the whole paper.
How Common Is It?
First of all, how common is multispecies hybridization? To quantify this phenomenon, I used records from the Serge Dumont Bird Hybrid Database for six bird orders with the highest incidence of hybridization: Anseriformes (waterfowl), Galliformes (wildfowl), Charadriiformes (waders, gulls and auks), Piciformes (woodpeckers), Apodiformes (hummingbirds and swifts) and Passeriformes (songbirds). This analysis revealed that most species hybridize with only one other species, but multispecies hybridization does occur frequently. In some bird orders, there are clear outliers (i.e. species that hybridize with many other species) such as Mallard (Anas platyrhynchos) Anseriformes, Common Pheasant (Phasianus colchicus) and European Herring Gull (Larus argentatus). These outliers have a large distribution range, providing ample opportunity to interact and potentially interbreed with several other species.

The incidence of multispecies hybridization in six bird orders.
Networks!
I have been advocating the use of phylogenetic networks for some time (see for example this paper in The Auk). In the current review paper, I introduce networks as a way to visualize patterns of hybridization. In its simplest form, you just connect all the species that are known to have produced hybrid offspring. Next, you could adjust the width or color of the connections based on other parameters, such as frequency, fertility, gene flow, … These hybrid networks are an important starting point for further exploration of multispecies hybridization.

A hybrid network for the Anseriformes. Dots represent species (colored according to different genera) while connections indicate that hybrid offspring have been observed.
Multispecies Introgression
Hybridization does not necessarily result in introgression (i.e. the exchange of genetic material from one (sub)species into the gene pool of another by means of hybridization and backcrossing). Although most statistical methods have been developed to quantify introgression between two hybridizing species, some approaches can be transferred to a multispecies setting. I will not cover all these methods here, but I will highlight a few interesting studies.
Clustering Methods
Model-based clustering methods, such as STRUCTURE and ADMIXTURE , are often used to visualize the genetic ancestry of individuals. This approach can be used to pinpoint individuals whose genomes show signs of ancestry from multiple sources. For example, Thies et al. (2018) uncovered a putative hybrid zone between three subspecies of the Common Ringed Plover (Charadrius hiaticula) based on STRUCTURE analyses. However, the output from these model-based clustering methods should not be taken at face value. Other processes can produce similar ancestry plots. That is why it is important to present the results of other analyses alongside these plots.

A STRUCTURE plot for the Common Ringed Plover. Three locations (in red box) point to a hybrid zone between three subspecies. Adapted from: Thies et al. (2018) Ardea.
The D-statistic and Phylogenetic Networks
The D-statistic was originally developed to infer gene flow between humans and Neanderthals. This approach was hidden deep in the supplementary material of a Science paper, but emerged as one of the most often used statistics to infer introgression. There are several bird studies that have applied the D-statistic in a multispecies setting. For instance, I used this method in my goose work to show that there is a lot of genetic exchange between several species, as exemplified by the phylogenetic network below. Did I mention that I like networks?

A phylogenetic network of the True Geese. From: Ottenburghs et al. (2017) BMC Evolutionary Biology
Modelling Introgression
There are numerous ways of modelling the demographic history of a species pair, ranging from Approximate Bayesian Computation (ABC) to diffusion-based models (e.g., DADI). Most of these models have not been applied in a multispecies setting yet. One notable exception concerns Isolation-with-Migration models that have been used to infer gene flow between three Acrocephalus warblers and between three Icterus orioles. New methods are being developed (as I witnessed at a recent Speciation Genomics workshop), but each of these approaches comes with its own assumptions (e.g., selective neutrality or small changes in allele frequency) and one should be aware of these when inferring patterns of gene flow.

An Isolation-with-migration model for three oriole species. The circles indicate effective population sizes and the arrows show the amount of gene flow. Adapted from: Jacobsen & Omland (2012) Ecology and Evolution
Adaptive Introgression
In his book on introgressive hybridization, Edgar Anderson (1949) already stated that “raw material brought in by introgression must greatly exceed the new genes produced directly by mutation.” Indeed, adaptive introgression has been documented in several bird groups, such as Darwin’s finches and sparrows (see this blog post for more details). When multiple lineages are interbreeding, the adaptive alleles can flow in from multiple sources. This is nicely illustrated by a recent study on the diversification and domestication of the bovine genus Bos. Comparing the genomes of members of this genus – which includes taurine cattle, zebu, gayal, gaur, banteng, yak, wisent and bison – revealed complex patterns of introgression between several species. Interestingly, both gayal and bali cattle received genes from zebu cattle through introgressive hybridization. In both cases, the introgressed genes were related to a decrease in anxiety-related behaviour and could have played a role in the domestication of these animals. You can read the whole story on cow introgression here.
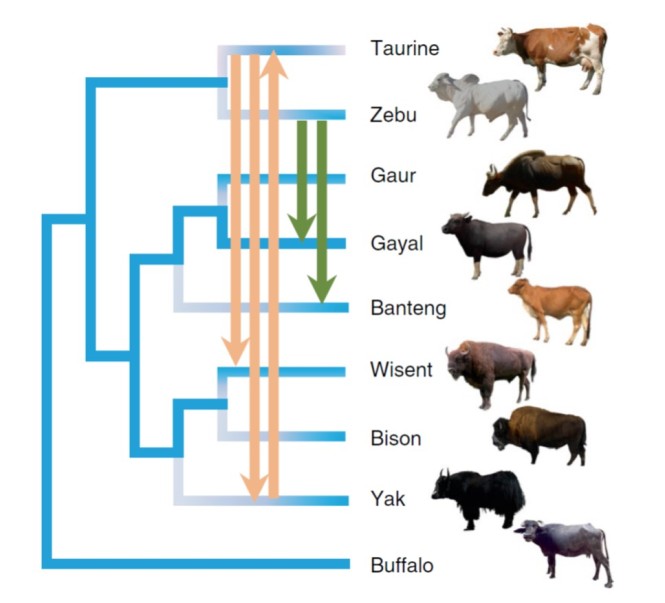
The complex evolutionary history of cows with multiple introgression events. From Wu et al. (2018) Nature Ecology & Evolution
Conclusions
To end this blog post, I will copy the conclusions of my review paper below. I hope more people will adopt this multispecies perspective on hybridization.
Hybridization is generally studied in the context of species pairs, although multiple species might be interbreeding. Hence, a multispecies perspective on hybridization is warranted. In birds, an animal group prone to hybridization, multispecies hybridization is common. However, hybridization does not necessarily result in introgression. A broad range of tools are available to infer interspecific gene flow. The majority of these tools can be transferred to a multispecies setting. Specifically, model-based approaches and phylogenetic networks are promising in the detection and characterization of multispecies introgression. At the moment, we know that introgression is relatively common across the avian Tree of Life, but we do not have an estimate of how often introgression is adaptive. In addition, when multiple species are interbreeding, the impact on the build-up of reproductive isolation, adaptation to novel environments and the architecture of genomic landscapes remains elusive. Studying hybridization between multiple species and applying new network approaches will lead to important insights into the history of life on this planet.
References
Ottenburghs, J. (2019) Multispecies Hybridization in Birds. Avian Research, 10:20.



































